CellCarta's Comprehensive Molecular Profiling Capabilities
As a leading Contract Research Organization (CRO), CellCarta serves as a trusted genomic service provider for pharma and biotech companies. Our comprehensive genomic solutions support pre-clinical and clinical studies. Our expertise spans various genomic technologies, providing accurate and reliable data.
Some of our capabilities including:
- Next-Generation Sequencing (NGS)
- Digital and quantitative PCR
- Single-cell sequencing
Our laboratories can support the handling and analysis of various sample types, and our analytical validation process is thorough and supported by partnerships with various reagent manufacturers.
All our workflows, including
- DNA/RNA extraction
- gene expression
- mutation analysis
- and data analysis
are performed under extensive quality control, providing essential genomic research support.
Using proprietary tools, our team can develop custom design qPCR/dPCR assays to further support your experiment.
You can rely on CellCarta as your genomics contract laboratory. We accompany you all the way to data analysis by offering a comprehensive Bio-IT pipeline and adapted molecular analysis software to deliver data analysis solutions that fit your needs.
Choose from our extensive list of genomic assays, fit for all phases of drug development from discovery and preclinical through clinical trials.
1- DNA/RNA Extraction
Some of our genomic solutions include methods for DNA and RNA extraction that take into consideration your downstream applications.
Our team has extensive experience in extracting various types of nucleic acids (genomic DNA, mRNA, cfDNA, cfRNA, miRNA and more) from numerous clinical sample types such as:
- whole blood, PBMCs, plasma, buffy coat
- cell lines
- FFPE
- FFT
- Fine-needle aspirate (FNA)
- and fresh tissues
We ensure quality for all of them with manual and automated methods. We also offer dual extraction possibilities where we simultaneously extract DNA and RNA separately from the same FFPE material.
Enrichment of specific tissue components or tumor cells by accurate manual microdissection prior to extraction is made possible through the combination of our digital pathology platform and our in-house, board-certified pathologists.
As a comprehensive genomics contract laboratory, our genomic research support team also specializes in developing exploratory extraction methods for novel conditions, uncommon tissue types, or complex multi-omics inquiries.
Our expertise goes beyond extraction, encompassing upstream kitting and logistics as well as downstream assay development, ensuring that we navigate your extraction method with all factors in mind.
Contact us for more information about our DNA/RNA extraction services!
2- Gene Expression Analysis (qPCR, dPCR)
Gene expression analysis is performed with sensitivity and specificity using PCR-based methods aligned with your study needs.
We support any qPCR or dPCR clinical applications with off-the-shelf or custom assays, utilizing technologies such as QuantStudio, Rotorgene, cobas systems, QIAcuity, and Bio-Rad QX200.
Exploratory assays are supported on QuantStudio, Lightcycler systems, QIAcuity, or Bio–Rad QX200, providing robust genomic research support for your projects.
Learn more about our digital PCR capabilities
Learn more about our quantitative PCR capabilities
3- RNA Profiling
CellCarta, your genomic service provider offers comprehensive RNA profiling information using next-generation sequencing (NGS) with platforms such as NovaSeq X Plus, NovaSeq 6000, and NextSeq 550Dx for various sequencing project sizes.
For a more targeted analysis of specific genes, RT-qPCR and the NanostringTM nCounter® system are offered to accurately quantify panels of targets of interest.
As a 10x Genomics certified service provider, we also provide single-cell RNA sequencing services using the Chromium X platform, facilitating your genomic research outsourcing needs.
Learn more about our RNA sequencing services
4- Mutation Analysis
At CellCarta, DNA mutation detections are performed using well-established single target mutation assays on different PCR platforms (qPCR, dPCR) as well as NGS analysis protocols.
We can detect both single and multiple target mutations, follow regulatory requirements, and perform custom design of sensitive mutation detection assays. We utilize NovaSeq X Plus, NovaSeq6000, NextSeq 500Dx, and MiSeq technologies with validated or customized panels, reporting identified variants and their clinical relevance.
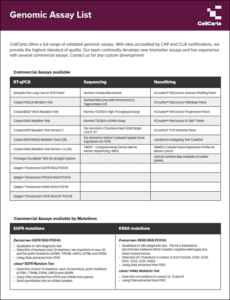
In addition to readily available targeted sequencing panels used to detect single nucleotide variants (SNV), small insertions, deletions, copy number variations (CNVs), and tumor mutational burden in specific genes frequently mutated in solid tumors, we also offer comprehensive panels:
- Pan Lung Cancer qPCR Panel by AmoyDx: Detects 167 mutations and fusion events in 11 target genes specific to lung cancer from FFPE samples.
- Aspyre-Lung by Biofidelity: Covers 11 NCCN genes including 114 variants (77 DNA mutations, 36 RNA fusions, 1 exon skipping event) from DNA and RNA extracted from FFPE, and cfDNA and cfRNA from blood samples.
- TruSight Oncology 500 (TSO500) Panel by Illumina: Detects >500 solid tumor biomarkers relevant to immuno-oncology, including MSI and TMB, using NovaSeq X Plus, NovaSeq 6000, and NextSeq 550Dx on both DNA/cfDNA FFPE and plasma samples.
- TSO Comprehensive assay by Illumina: An in vitro diagnostics test detecting variants in 517 genes (SNVs, MNVs, insertions, deletions, gene amplifications from DNA; gene fusions, splice variants from RNA) in FFPE tumor tissues with NextSeq 550Dx. It also reports TMB score and MSI status and is validated for patient management in EU and US.
- TruSight Oncology 500 ctDNA V2 by Illumina: A pan-cancer panel targeting 523 genes to assess all main variant classes (SNV, MNV, indels, CNV, gene rearrangement) in ctDNA from blood plasma.
- OncoReveal CDx by Pillar: A pan-cancer solid tumor panel (NSCLC, CRC, Breast, Ovarian) with 22 genes and 103 amplicons for FFPE DNA.
These offerings provide essential genomic research support for drug development and clinical trials. Contact us for more information about our Mutation Analysis services!
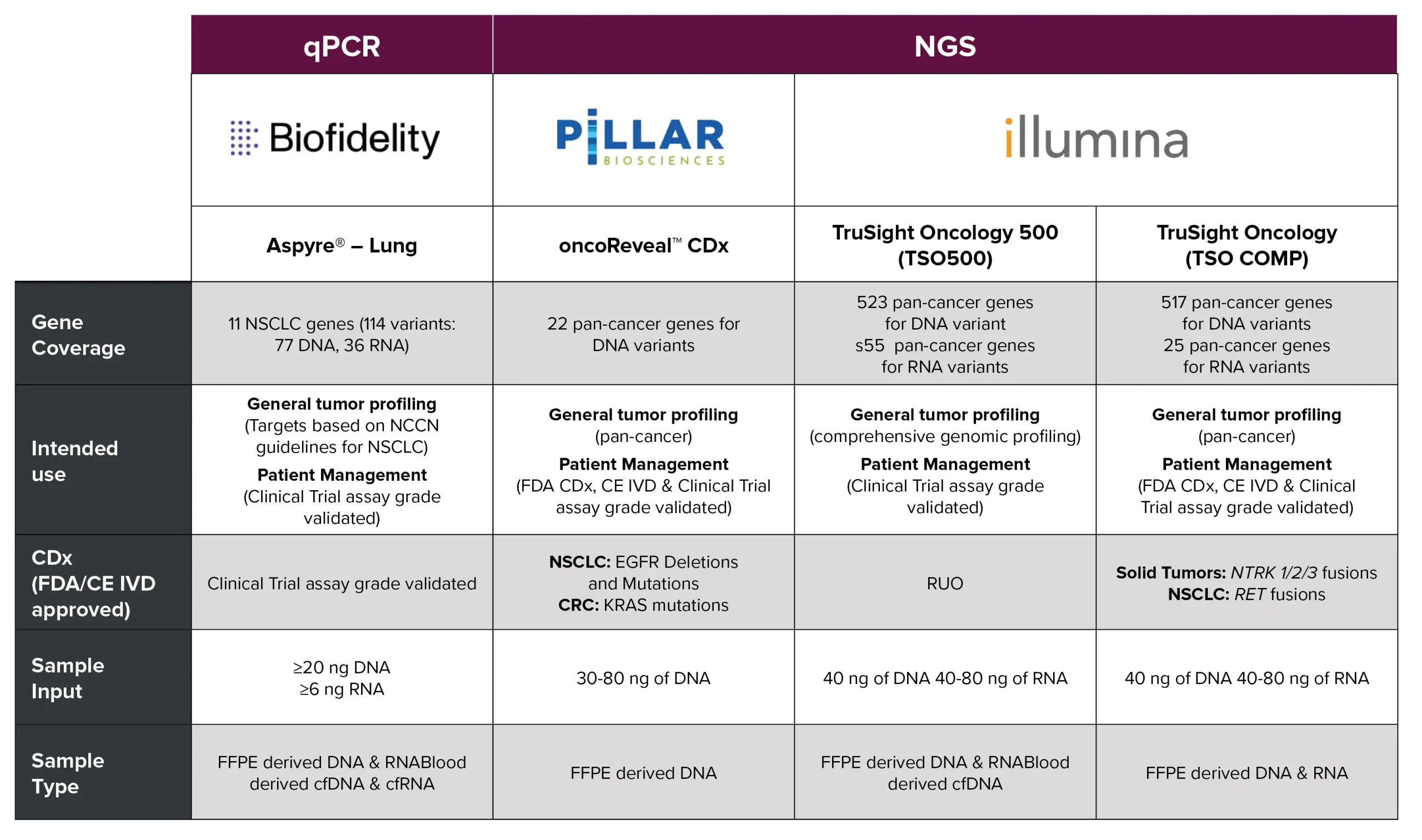
5- Fusion Detection
For RNA fusion detection, we utilize targeted NGS, RNASeq, qPCR, and digital PCR. Validated panels are used to detect cancer-specific fusions and point mutations, some of which do not require knowledge of specific break points.
Ready-to-use panels include:
- Pan Lung Cancer qPCR Panel by AmoyDx (real-time PCR): 11 target genes for lung samples
- TSO500 Panel by Illumina (NGS): 523 DNA and 55 RNA targets
- Customization for specific fusions and point mutations is also available, offering flexible genomic solutions for biotech and pharma.
Contact us for more information about our Fusion Detection services!
6- Microsatellite instability (MSI) Assays
Microsatellite instabilities (MSI) result from the systematic accumulation of deletions/insertions in short repetitive DNA sequences in tumor cells due to a deficient mismatch repair (MMR) system.
MSI occurs in many colorectal cancers and is clinically useful in identifying patients with HNPCC (Lynch Syndrome) caused by germline mutations of MMR genes.
The MSI status may also predict a patient’s response to certain chemotherapies. More recently, it has been used as a biomarker for immunotherapeutic response, making the MSI status an increasingly relevant tool in genetic and immuno-oncology research.
At CellCarta, we use panels consisting of quasimonomorphic mononucleotide repeats to determine MSI status.
- The PromegaTM MSI assay v1.2 provides a high throughput solution for multiplex amplification of five repeat markers (BAT-25, BAT-26, NR-21, NR-24 and MONO-27) in the DNA to determine MSI status.
- The TSO500 Panel by Illumina can also be used to determine the MSI status.
Our RNA IO data analysis workflow is performed on bulk RNAseq data from FFPE tumor samples and can be used to investigate both MSI and TMB status, as well as predict immune checkpoint inhibition response.
Contact us for more information about our microsatellite instability services!
7- Single-Cell Profiling and Spatial Gene Expression (10XGenomics – Chromium, Visium)
Using the Chromium system from 10XGenomics, CellCarta can provide single-cell RNASeq profiling.
The heterogeneity of samples normally masked in bulk RNA sequencing analysis can be captured through this method by profiling thousands of individual cells.
The assay is customizable to detect the whole transcriptome, the expression of targeted genes, specific surface proteins, and even single guide RNA (sgRNA) from CRISPR screen.
With the Visium system from 10X Genomics and GeoMx DSP from Nanostring-our team can provide a spatial map of the whole transcriptome within the morphological context of the tissue.
Spatial gene expression data is key in identifying spatiotemporal gene expression patterns and can offer great insight on the tumor and its immune microenvironment.
Contact us to explore how our specialized genomic services and research support can meet your specific project needs!
Why CellCarta a your Genomics CRO?
Genomics plays a crucial role in areas like oncology, immunology, and rare disease research. As a trusted partner in genomic research delivering quality results, we accelerate discoveries in these fields.
At CellCarta, we are ready to partner with you and apply our expertise as a leading Contract Research Organization to help accelerate your genomic research.