NanoString: Multiplexed Assessment of Gene Expression and Copy Number Variation in Clinical Samples
NanoString Technology is Digital and Automated, Reproducible and Precise
The NanoStringTM technology is used to capture and count specific DNA (CNVs, SNVs, IN-Dels), RNA (mRNA, miRNA, fusions), or proteins (including post-translational modifications) using a system of color-coded probes.
Its advantages over existing platforms include direct measurement of target levels without enzymatic reactions or amplification bias, sensitivity coupled with high multiplex capability, and digital readout.
The NanoStringTM – nCounter® gene expression system is more sensitive than microarrays and similar in sensitivity and accuracy to real-time PCR.
Given it can measure even low abundance mRNA in small samples, it is well suited to obtain expression profiles and specific signatures in clinical samples.
CellCarta offers a sensitive and reproducible gene expression analysis of up to 800 genes in a single reaction using the NanoStringTM technology.
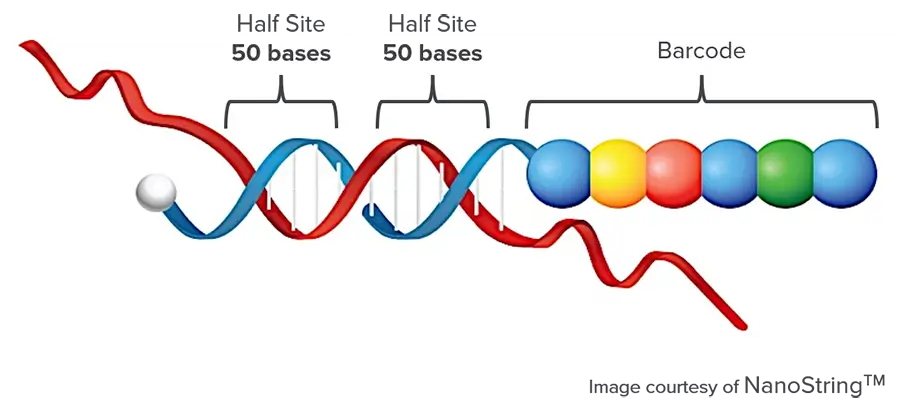
Unique Code Assigned to Each Gene
Digitally count up to ~1 million molecules per sampling area using two target-specific probes.
A capture probe (~50 base pairs) is complementary to the target sequence and coupled to a biotin tag, and a color-coded probe (~50 base pairs) also complementary to the target sequence, but coupled to a color-coded tag providing the detection signal.
This colored tag consists of a series of complementary in vitro transcribed RNA segments each labeled with a specific fluorophore.
A unique code is created from their order of presentation and is used to identify the target.
Once the target is bound by both probes, it is isolated using a streptavidin-coated surface, immobilized in an elongated state using an electric field, and imaged.
The specific order of the fluorescent signal allows for the identification and measurement of target.
NanoString System's Capability in Multiplexing Samples & Targets
The NanoStringTM system is capable of a high degree of multiplexing, performing gene expression for up to 800 gene targets in one single nCounter® experiment.
Ideal for providing gene expression signatures in clinical samples, it generally requires 100ng of total RNA (in a volume of 5-10 µl) for most gene expression panels.
The required number of FFPE glass slides depends on the biopsy types and cellularity. The following are general guidelines:
- Large resection specimens with high cellularity
– 2 to 4 sections of 5 µm - Small biopsies or specimens with low cellularity
– 4 to 10 sections of 5 µm
The simple protocol leads to a sensitive, precise, and quantitative count. The digital data output includes a list of genes investigated and the number of times they were observed in the samples.
Different data analysis packages are offered to ease data interpretation, including box plots by treatment group, scatter plots of all genes, specific advanced analysis, and clustering and heat maps.
Available Panels for NanoString Technology
Readily panels are offered and can be customized based on needs. Of interest is the Pan Cancer 770-plex panel used in immunotherapy clinical trials to investigate key cancer driver pathways.
- nCounter® Gene Expression Panels
- PanCancer IO 360TM Gene Expression Panel
(750 cancer-related genes) - PanCancer Immune Profiling Panel
(770 immune profiling genes) - PanCancer Progression Panel
(770 genes involved in cancer progression) - PanCancer Pathway Panel
(770 genes for essential cancer pathways)
- PanCancer IO 360TM Gene Expression Panel
- nCounter®-based Lymphoma Subtyping Test (LST)
for Cell-of-Origin (COO) - PAM50 Breast Cancer Subtyping Assay