Comprehensive cytometry analysis software
CellEngine Software
CellEngine powers CellCarta’s global flow cytometry analysis services, allowing us to deliver high-quality data to clients in record time. You, too, can supercharge your data analysis by leveraging CellEngine as a software-as-a-service (SaaS) to analyze your own data.
Learn more about how we use CellEngine at CellCarta, or read below to learn more about the features of the SaaS offering.
Unmatched Performance
Speed through analysis, whether you have just a few tubes, dozens of 384-well plates, or multi-gigabyte FCS files.
The largest experiment analyzed in CellEngine to date included 30,000 FCS files. With that capacity, CellEngine is uniquely able to analyze large drug screens or entire longitudinal studies spanning months or years in a single, coherent view.
Easy Autogating
CellEngine’s supervised autogating tool uses machine learning to automatically tailor your gates according to your hierarchy.
Gate positions can be adjusted in seconds by using a small set of manually gated files from your dataset. The tool mimics your gate placement decisions, allowing autogating to seamlessly supplement existing manual gating steps. Save time and allow analysts to focus on data interpretation.
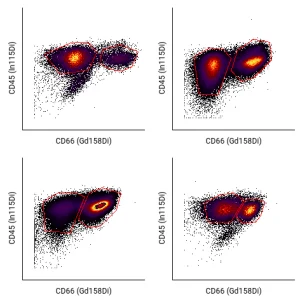
Learn more about easy and customized autogating with Cellengine
Advanced Gating Features
CellEngine has built-in tools to make gating easy and fast, no matter how complex your gating hierarchy. Performing multifunctional cytokine analysis? Combination gates will reveal the profile of every cell. Trying to set a gate position based on an unstimulated control? Use percentile gating to place vertices at a specific percentile.
Advanced Charting and Visualizations
Create bar and line charts, box plots, annotated and clustered heatmaps, flow plot overlays, gating hierarchies, and dose-response curves.
Use batched illustrations to create reports that automatically update as your study progresses.
Analyze replicates and assess long-term instrument and assay performance with Levey-Jennings plots and the ability to view the staining profiles underlying line chart data.
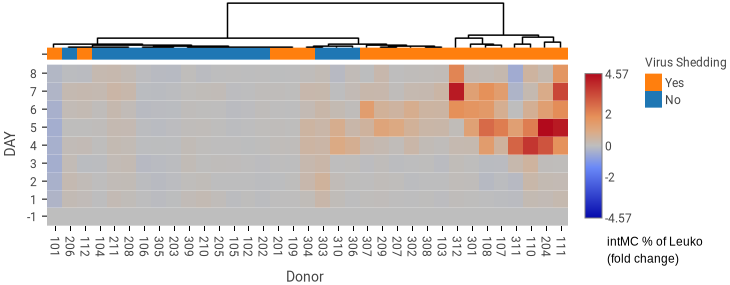
Rahil Z et al. J Clin Invest 2020;130(11):5800-5816.
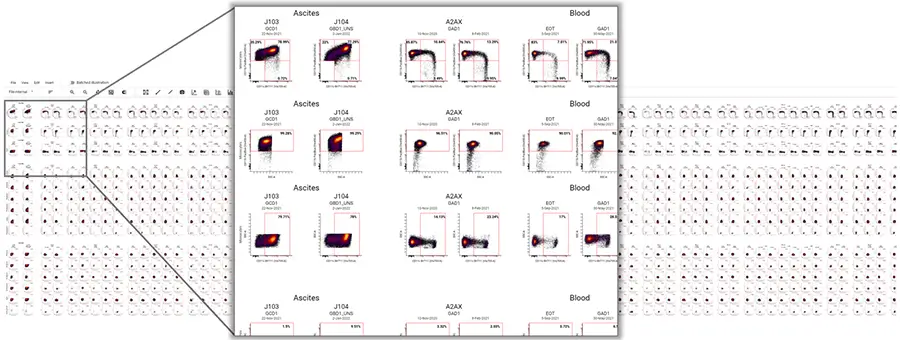
Metadata-Driven Analysis
Start by adding sample metadata, such as timepoint, donor, dosage, and treatment, using a familiar spreadsheet interface. Then, use that metadata to systematically gate your data, automatically lay out your visualizations, or filter your dataset down to a subset of interest. This approach reduces human error and saves time over manually organizing and summarizing data.
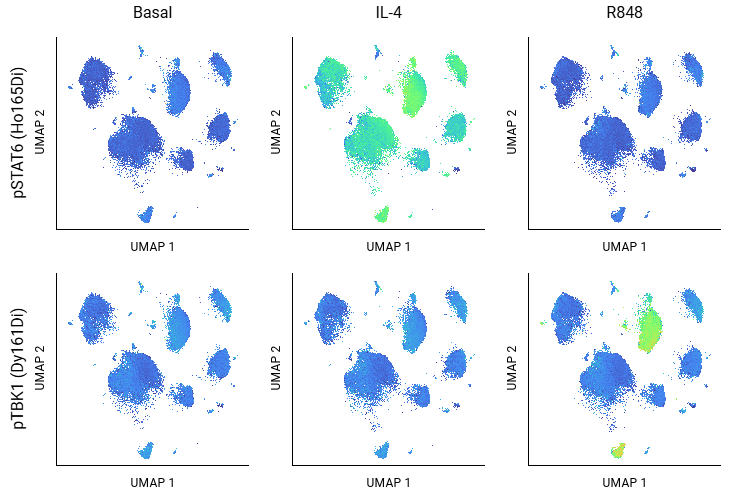
Fast Algorithms
Easily run dimensionality reduction and clustering algorithms, including UMAP and FlowSOM. CellEngine’s optimized versions can analyze tens to hundreds of millions of cells in minutes, not hours. The results can be subsequently visualized without leaving CellEngine.
Interoperable and Compatible
Easily import experiments from other widely used cytometry software like BD FACSDivaTM and Cytobank.
CellEngine is compatible with FCS files from more than 45 different cytometers and over a dozen manufacturers. Analyses and results can be exported in a variety of standard formats.
Safe and Secure
By using geo-redundant storage, we ensure the protection of your data against natural disasters and hardware failures. All data is encrypted in-flight and at-rest using strong ciphers. Learn more about CellEngine’s safety and security.
Cloud-Based
All of your experiments are organized in one place, accessible from any Internet-connected computer. By default, only you can access your data, but you can easily share it with coworkers and collaborators.
Advanced API
CellEngine offers a fully featured API. Integrate CellEngine with your LIMS, ELN, or EMR and conduct advanced analyses as part of bioinformatics pipelines.
We provide and support API toolkits for R and Python.
Regulatory Compliance
CellEngine provides all the features required for use in 21 CFR 11-compliant environments.
CellCarta continuously validates the software with thousands of automated tests to ensure consistent results and performance
Cited in Peer-Reviewed Publications
Publications in Cell:
Rahim MK et al. Cell 2023;186(6):1127-1143.
Publication in The Journal of Quantitative Cell Science:
Fallahzadeh R et al. Cytometry 2021;12(1):2338. doi:10.1002/cyto.a.24709.
Publication in Science Advances:
Mayer A et al. Sci Adv. 2023;9(3): doi:10.1126/sciadv.add1166
Publication in Nature Medicine:
Padrón J. L et al. Nat Med. 2022;28:1167-1177; doi:10.1038/s41591-022-01829-9.
Publication in Clinical Cancer Research:
Mettu B. N et al. Clin Cancer Res. 2022;28(5):882-892; doi:10.1158/1078-0432.CCR-21-2780.
Publication in The Journal of Clinical Investigation (JCI):
Rahil Z et al. JCI 2020;130(11):5800-5816.