August 18, 2025
Making the most of limited tissue samples
In clinical trials, patient tissue samples are limited, so extracting the most valuable data you can from each slide is critical.
Combining immunohistochemistry (IHC) and RNA sequencing (RNAseq) on the same sample offers a way to increase the insight gained from limited material. However, while many teams are aware of the potential to extract both spatial and molecular data from a single slide, they often exclude RNAseq due to tissue and budget constraints, viewing it as purely exploratory rather than essential for clinical decision-making.
But, with the right approach, RNAseq can be added alongside IHC to unlock molecular insights that extend the value of each tissue sample, generating richer, more connected data.
Extracting deeper insights with IHC and RNAseq combined
While IHC provides spatial detail and protein-level expression, RNAseq adds molecular depth, capturing bulk gene expression across a sample to reveal underlying transcriptional activity and mutational burden, supporting a wide range of research applications.
When combined with IHC data from the same sample, RNAseq can offer deeper insight than IHC alone, allowing for:
- A stronger understanding of a drug’s mechanism of action
- Better biomarker discovery and refinement over time by bridging discovery to research and implementation
- Richer, comparable data across time points in longitudinal studies
- Correlation of transcript abundance with protein localization and expression in specific cell types
- Dissection of cellular heterogeneity within a tissue sample
- A fuller picture of the tumor-immune landscape, which is particularly valuable for immunotherapy studies
IHC and RNAseq can also be scaled across large sample volumes, making the combined approach suitable for large-scale clinical studies.
Get in touch to find out how we can support combined IHC and RNAseq analysis
Case study: RNAseq for predicting immune checkpoint inhibition response
In immuno-oncology, predicting patient response to immune checkpoint inhibitors (ICI) presents a major challenge. While several features have been shown to correlate with ICI response—e.g., tumor mutational burden (TMB), microsatellite instability (MSI), immune gene expression signatures, and tumor-infiltrating lymphocytes (TILs)—measuring them typically requires multiple different omics techniques.
However, combining multiple techniques is costly and time-consuming, and—most importantly—consumes precious patient material that is often limited and irreplaceable. RNAseq, on the other hand, can overcome these challenges by serving as an all-in-one omics solution, providing comprehensive molecular information from a single experiment.
Through a specially developed suite of data processing and analysis pipelines, CellCarta has established a method of extracting all four key features from RNAseq of tumor samples (Figure 1).
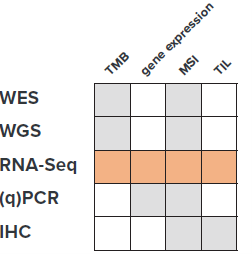
Figure 1: Summary of established features associated with ICI responses and the different techniques used to measure them. WES: whole-exome sequencing, WGS: whole-genome sequencing, RNA-Seq: RNA sequencing, qPCR: quantitative PCR, IHC: immunohistochemistry.
Not only does this method reduce the cost and time associated with analysis, but it also offers the possibility to improve ICI response prediction by integrating features into a single model (Figure 2). For more information on how our RNAseq technique improves response predictions, check out our white paper.
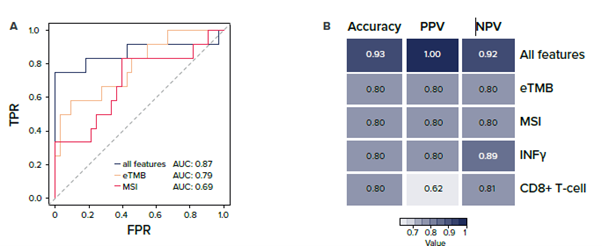
Figure 2: Improved response prediction when integrating all features compared to individual features. A) Logistic regression model incorporating all features (eTMB, MSI, CD8+ T-cell, M1 macrophage, IFNγ signature, and CTL signature; AUC = 0.87) vs. eTMB (AUC = 0.79) and MSI (AUC = 0.69). Higher AUC value for all features compared to eTMB or MSI scoring alone. Prediction performance is demonstrated by a ROC curve and area under the ROC curve (AUC). B) Performance metrics for each model. AUC: area under the ROC curve, TPR: true positive rate, FPR: false positive rate, PPV: positive predictive value, NPV: negative predictive value.
By enabling comprehensive profiling of the tumor and tumor microenvironment from a single sample, CellCarta’s RNAseq workflow makes it easier to generate clinically relevant insights without adding complexity or consuming more tissue. Combined with IHC to add vital spatial and protein-level context, this approach delivers a deeper, more connected picture of tumor biology from limited material, supporting more informed predictive biomarker development and patient selection decisions.
Enable integrated tissue profiling with confidence
CellCarta supports clinical and translational research teams in implementing IHC and RNAseq workflows that are both efficient and reproducible. Our infrastructure and expertise help ensure high-quality results from limited tissue across sites and studies. We offer:
- Access to all major sequencing platforms, enabling flexibility across study designs
- Off-the-shelf and customized assay options, depending on your biomarker strategy
- Global reach, with standardized SOPs and coordinated operations across trial sites
- Expert support for all samples, enabled by in-house pre-analytical services
Whether you’re integrating IHC and RNAseq for the first time or scaling up for a multi-site study, CellCarta is equipped to help you get more from every sample.
Want to explore IHC and RNAseq integration in your next project? Contact our experts
You might also be interested by
CellTalk Blog
Leveraging Preclinical Genomic Insights for Successful Therapeutic Development
August 18, 2025
Genomics
More infoCellTalk Blog
Immortalizing Your Samples: The Power of Dual Extraction in Pre-Analytical Planning
August 18, 2025
Genomics
More infoWeb News
Biofidelity and CellCarta partner to deploy Aspyre® Lung in global clinical trials
January 9, 2025
Genomics
More infoCellTalk Blog
How can HLA typing drive better immunotherapy development and selection?
December 6, 2023
Genomics
More info