November 5, 2025
RNA sequencing (RNA-seq) is a critical tool in biomarker discovery and translational research. Its accuracy and efficiency depend heavily on the quality of library preparation, which can affect everything from the number of genes detected to the accuracy and reproducibility of gene abundance estimates. An inadequate workflow at this stage can lead to long preparation times, unreliable data, and, at worst, failed samples.
When working with valuable clinical material, it is therefore vital to use the most reliable and efficient RNA library preparation method available. At CellCarta, we continuously refine our workflows to deliver the highest-quality data. As part of this commitment, we recently evaluated the Watchmaker Genomics (WMG) RNA-sequencing workflows as an alternative to standard capture RNA-sequencing methods. The Watchmaker workflow reduces preparation time from 16 hours to only 4 hours, all while improving data quality, data yield, and reproducibility.
Below, we highlight results from our validation study and the advantages Watchmaker Genomics can bring for your RNA-seq workflows.
Comparing Watchmaker Genomics with standard capture RNA-sequencing
In our validation study, the Watchmaker RNA library prep with Polaris® Depletion was benchmarked directly against the standard RNA capture method. The results showed consistent improvements across multiple performance measures, including duplication rates, mapping rates, and gene detection.
Lower duplication rates
High duplication rates lead to wasted sequencing capacity and can skew gene abundance estimates. With Watchmaker RNA library prep with Polaris Depletion, duplication rates were significantly reduced, and uniquely mapped reads were significantly increased compared to the standard method (Figure 1), resulting in cleaner data, a more efficient use of sequencing resources, and allowing for more reliable biological insights.
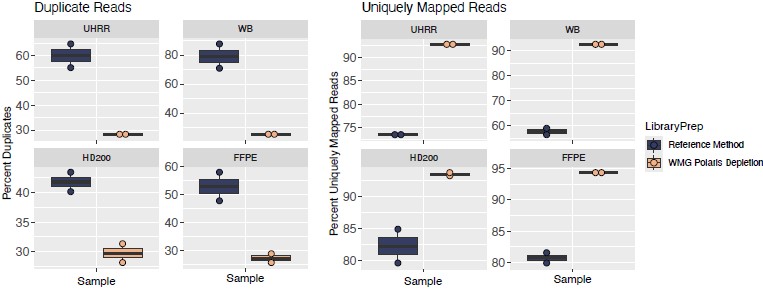
Figure 1: Watchmaker demonstrates a significant reduction in PCR duplication rates and a higher fraction of uniquely mapped reads compared to the standard RNA capture method, independent of sample type. UHRR: universal human reference RNA, WB: whole blood, HD200: Horizon Discovery reference sample, FFPE: formalin-fixed paraffin-embedded, WGM: Watchmaker Genomics.
Efficient depletion of rRNA and globin
Another area of improvement with Watchmaker RNA library prep with Polaris Depletion was the removal of unwanted RNA species. Poor removal of ribosomal RNA (rRNA) and globin RNA results in fewer reads that map to the biologically informative portion of the transcriptome, thereby negatively impacting sequencing efficiency. In our validation, Watchmaker RNA library prep with Polaris Depletion consistently reduced both rRNA and globin reads in both formalin-fixed paraffin-embedded (FFPE) samples, and whole blood (WB), compared to the standard RNA capture method.
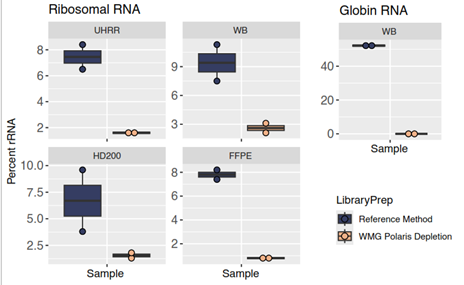
Figure 2: The Watchmaker workflow generated fewer rRNA reads and a reduction in globin reads compared to the standard RNA-seq method.
More detected genes
Watchmaker RNA library prep with Polaris Depletion also enabled the detection of 30% more genes across sample types compared with the standard capture method (Figure 3), reflecting the higher proportion of informative reads and allowing deeper coverage of the transcriptome. For researchers, this means richer datasets, stronger biomarker discovery potential, and more confidence in downstream analyses.
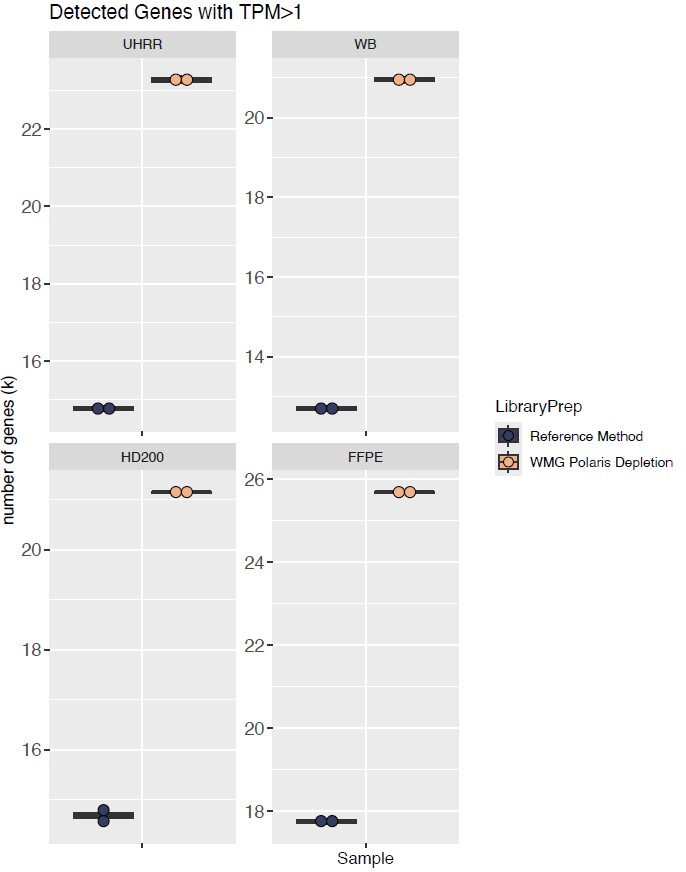
Figure 3: Watchmaker libraries consistently detected more genes across sample types compared with the standard RNA capture method. TPM: transcripts per million.
Maximize the value of every sample
Our validation confirmed that the Watchmaker Genomics RNA-seq workflow delivers consistent improvements in duplication rates, gene detection, and reproducibility compared with standard RNA capture methods. For researchers, this means cleaner libraries, richer transcriptome coverage, and greater confidence that they can get meaningful insights from their samples.
Contact us to learn more about how CellCarta can apply the Watchmaker library prep workflow to support your RNA-seq needs.
About the author:

Pieter Mestdagh is a genomics expert at CellCarta. With a PhD degree in biomedical sciences, and a broad expertise gained through his work in academia and industry, Pieter has broad experience with nucleic acid quantification methods to support DNA and RNA analysis in clinical trials.
You might also be interested by
CellTalk Blog
How to Enhance Clinical Trial Patient Enrollment with Molecular Profiling
August 18, 2025
Genomics
More infoPosters
A Quantitative Mass Spectrometry Workflow for Highly Multiplexed Measurement of Immunomodulatory Proteins to Support Immunotherapy Clinical Trials
October 13, 2023
Genomics
More infoCase Studies & Whitepapers
Accurately Predict Immune Checkpoint Inhibition Response Using Only RNA-Seq Data
June 27, 2023
Genomics
More info