January 31, 2023

Analyzing data is one of the most challenging and rewarding parts of an experiment.
CellEngine by CellCarta leverages metadata during every step of cytometry analysis, from initial gating to final figure details, increasing the speed and depth of analysis possibilities.
Add Any Type of Metadata for Comprehensive Cytometry Analysis
Metadata can be entered directly or imported from a spreadsheet. In addition, there are tools for adding information that was included in a filename or FCS header.
Any information can be added, but common categories include:
- Assay – the preparation conditions, run date, cytometer, or researcher who ran the sample
- Sample – treatment condition or concentration, timepoint, or results from other assays
- Source – cell line, disease information, donor ID, or demographic data
Data validation rules can be added to restrict values that are accepted for each variable, creating a robust defense against typos, miscopied data, or inconsistencies such as “IL-2” versus “IL2.”
Use Metadata to Select and Organize Files
Everywhere files are selected in CellEngine, they can be sorted and filtered by metadata, including when gating, creating a figure, using the built-in algorithms for dimensionality reduction and clustering, or exporting statistics.
This allows for straightforward selection of any group of files without risk of human error.
Generate Accurate, Insightful Figures Using Metadata
Metadata can be used to organize data into figures, acting as axes or data series.
This allows for easy creation of a wide number of figures – for example,
- a line graph organized by timepoint
- a set of histograms overlaid by treatment
- a dose-response curve using concentration
- a bar graph comparing populations normalized to a control condition.
Plots with multiple files can be further filtered on any metadata value, for example creating a figure using only samples from healthy donors.
Furthermore, batching can iterate through selected values of one or more metadata variables to automatically generate a series of figures.
If analysis is started before an experiment is complete, such as early during a months-long clinical trial, figures can be set to automatically update when new files are uploaded.
This allows users to begin work between timepoints or replicates, decreasing time between experiment completion and figure finalization.
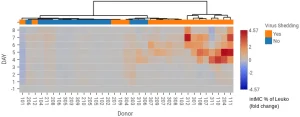
Figure 1: The ability to add additional metadata to heatmaps reveals a correlation between increased intermediate monocytes and shedding of virus during influenza infection. Data from Rahil Z et al. J Clin Invest 2020;130(11):5800-5816.
Accelerate Metadata-driven Cytometry Analysis with CellEngine
CellEngine’s metadata features are contained in a highly optimized, cloud-based analysis platform.
Together, users can iterate on their analysis and pursue hypotheses rapidly as questions emerge.
In addition, CellEngine has many other features designed to improve flow cytometry analysis, particularly for large and complex experiments.
Sign up for a free two-month trial or contact us for a live demo.
Explore In-Depth Features of CellEngine Software from our Expert
About the author:

Anita Ray is a technical application specialist with CellEngine. She has ten years of experience in cytometry and worked in translational immuno-oncology prior to joining the CellEngine team.
You might also be interested by
CellTalk Blog
ELISpot vs. ICS: Optimizing Immune Monitoring in Clinical Trials with the Right Functional Assay
November 19, 2024
Immune Monitoring
More infoCellTalk Blog
Measure Target Engagement With Receptor Occupancy Assays by Flow Cytometry
September 17, 2024
Immune Monitoring
More infoBrochures & Infographics
High Performance Technologies for Cytokine Measurement
July 29, 2024
Immune Monitoring
More info